English
Predict tuberculosis resistance to anti-TB drugs
Deeplex® Myc-TB is an all-in-one test for species-level identification, genotyping and prediction of antibiotic resistance in Mycobacterium tuberculosis complex strains.
Exome sequencing
Exome sequencing allows targeting the genome coding regions. GenoScreen offers Whole Exome Sequencing (WES) services through the NimbleGen SeqCap EZ System (Roche).
Sanger Sequencing
GenoScreen's historical activity since 2001, Sanger sequencing is used in all fields and for a wide range of applications. It is the standard technique for sequencing DNA accurately and robustly. On our dedicated platform, we offer routine sequencing and, in addition, customised services with various levels of intervention and analysis.
Our Sanger sequencing platform
Equipped with ABI 3730XL capillary sequencers, our platform meets your one-off and regular sequencing needs from tubes, strips or 96-well plates containing your PCR products or vectors.
Molecular barcoding
Molecular barcoding is a technique for identifying and genetically characterising a sample by targeting a ubiquitous gene, often mitochondrial or chloroplast.
Applications:
- Identification of a species from an isolated sample (microorganism, animal or plant species)
- Comparison of several samples from the same species or closely related species

Services offered by GenoScreen
- DNA extraction from any type of matrix (optional)
- PCR amplification, sequencing and assignment of samples to databases (NCBI, BOLD, etc.)
- Construction of phylogenetic trees
- MLST typing and whole genome sequencing (WGS) to characterise the genome
- Solutions for complex matrices via metabarcoding
Discover our services in molecular barcoding
Additional Sanger services
Additional Sanger services
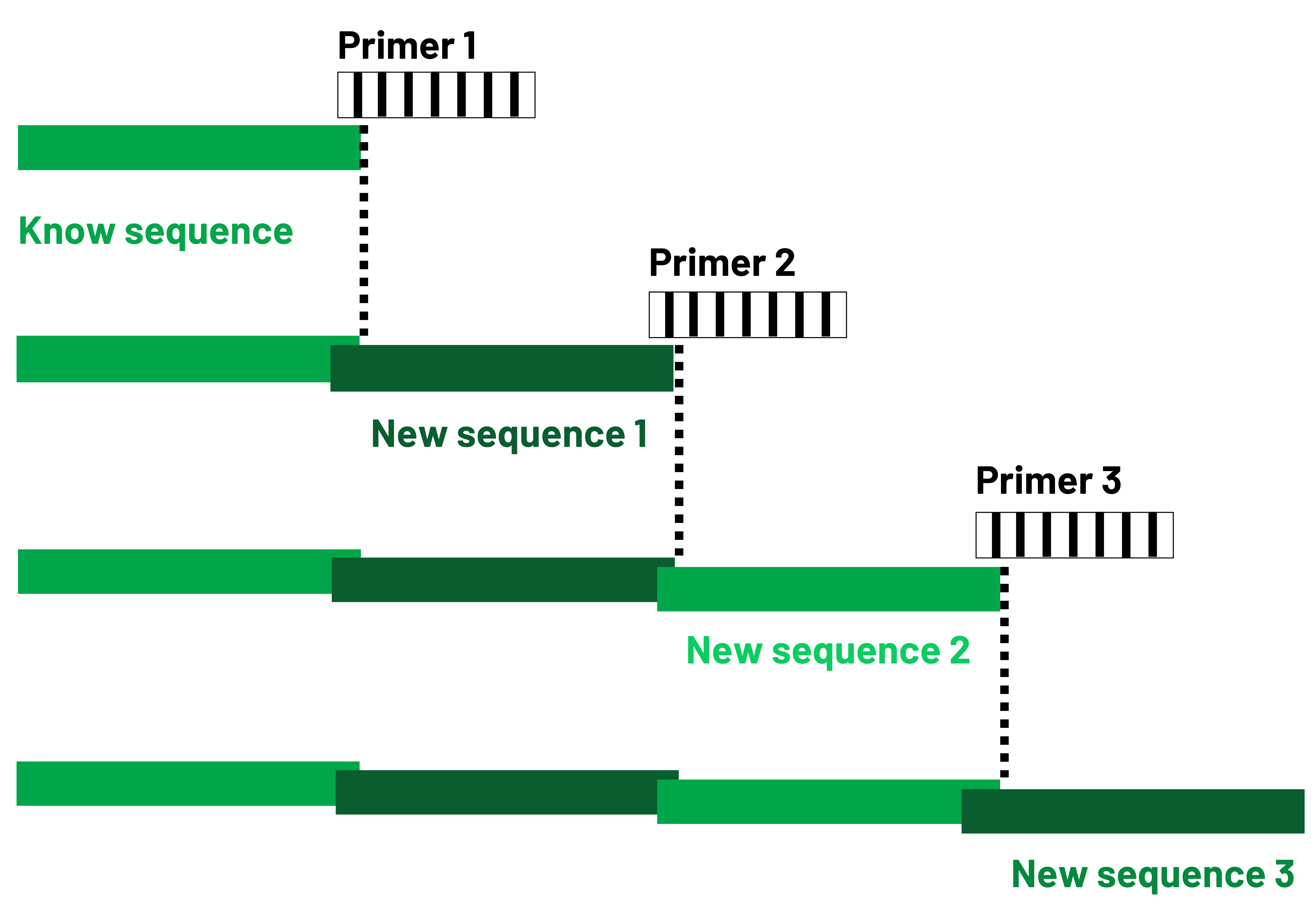
Primer walking
This sequencing method is ideal for:
Principle
The use of several primers makes it possible to obtain overlapping sequences leading to the reconstitution of the complete product. A universal primer is used to sequence an insert, obtaining a fragment of the vector sequence and the beginning of the insert.
A specific primer can then be designed on the sequence obtained and used to obtain the rest of the sequence. If the sequence of interest is known, all the sequencing reactions can be carried out simultaneously. This service can be performed either in single or double reading.
This email address is being protected from spambots. You need JavaScript enabled to view it.
Mutation detection
Sanger sequencing can be adopted to verify
Principle
Among other applications, the mutation detection is used to monitor TILLING (Targeting Induced Local Lesions in Genomes) in plants, or to verify mutations in genes of interest.
We can design specific primers to target a mutation and compare it to a reference sequence provided for verification.
If several mutations/positions are to be verified in the same genome region, we can also produce a detailed variance report compared with a reference gene.
Identification - Molecular barcoding
Molecular barcoding (DNA barcoding) is a molecular taxonomy technique used to identify and genetically characterise a sample from a ubiquitous gene, generally belonging to the ribosomal, mitochondrial or chloroplast genome (COI, 16S, rbcL, etc.).
DNA barcoding applications
DNA barcoding can be used to:
- Identify species from an isolated sample
- Compare several samples from the same species or closely related species (phylogeny)
This email address is being protected from spambots. You need JavaScript enabled to view it.
Identification by DNA barcoding
GenoScreen offers a complete molecular barcoding service and can handle your samples from DNA extraction onwards.
- Identification of isolated micro-organisms
- Identification of animal or plant species
For complex inital matrices (e.g. assemblies of bacterial populations), GenoScreen develops metabarcoding solutions using NGS amplicon sequencing to obtain screenings of samples: soil fauna, intestinal flora, coral reefs, etc.
Sequencing
GenoScreen is a major international player providing sequencing and genomic analysis services.
The company’s expertise is based on an unique experience in genomic analyses, the use of cutting-edge technologies and the development and supply of exclusive bioinformatics tools and R&D services. GenoScreen offers a very broad range of sequencing services, from the most basic to the most advanced.
NGS - High-power genomics for the life sciences
Since 2008, GenoScreen offers a full range of next-generation sequencing (NGS) services in application to genomes, metagenomes and transcriptomes.
GenoScreen has cutting-edge technical facilities, giving you access to the latest technologies (Illumina, PacBio, etc.).
The GenoScreen's assets
- A full range of NGS services
- Expert analysis and advice on the design of each analysis
- Unique expertise in microbiology
- High-quality data
- Exclusive bioinformatics tools: assembly, annotation, SNP detection, etc.
- Guaranteed short delay time
This email address is being protected from spambots. You need JavaScript enabled to view it.
Subcategories
GenoScreen
Collaborative research - At the heart of international innovation in genomics
GenoScreen is a partner in collaborative research projects that bring together startups, multinationals and public-sector organizations. Our R&D teams provide their knowledge and expertise in the molecular microbiology of isolated agents and complex communities. This allowed our teams to develop our own projects for elaborating innovative products and services.
Our projects are designed to:
- Improve the diagnosis and management of acute/chronic diseases on humans and animals,
- Characterize and monitor microbial biodiversity, with applications in agronomy, agrifood and environment.
The common feature of these projects is the development of molecular tools for the characterization, monitoring and diagnosis of microbial communities. The key objective is to market simple analytical solutions and (ultimately) preventive, corrective or even therapeutic products based on microorganisms.