News
-
Staphyt and GenoScreen join forces to offer integrated solutions in genomics, regulatory studies and field trials
21/01/2026 — Staphyt, a leading contract research organization (CRO) and regulatory affairs consultant for agricultural experimentation and risk assessment, and GenoScreen, a pioneer and specialist in genomic and bioinformatics solutions, are pleased to announce the signing of a strategic collaboration agreement.
-
New EFSA Guidance for microorganisms (2025) : What changes and how GenoScreen help you stay fully compliant
Following its agenda to regulate the Food and Feed industry with up-to-date regulatory texts, the EFSA significantly raised the technical expectations for the characterisation and risk assessment of microorganisms used in food and feed. As of November 4th, 2025, dossiers that rely on outdated genomic analysis, incomplete resistance screening or legacy databases may no longer meet regulatory expectations.
-
How to scientifically and regulatory validate a probiotic or postbiotic Strain (EFSA, FDA)?
What are the requirements for marketing a probiotic or postbiotic strain? Developing supplements or functional foods containing a biotic strain involves much more than simply demonstrating their effectiveness. This process is essential for obtaining regulatory approvals from authorities such as the European Food Safety Authority (EFSA) and the U.S. Food and Drug Administration (FDA) :
Focus
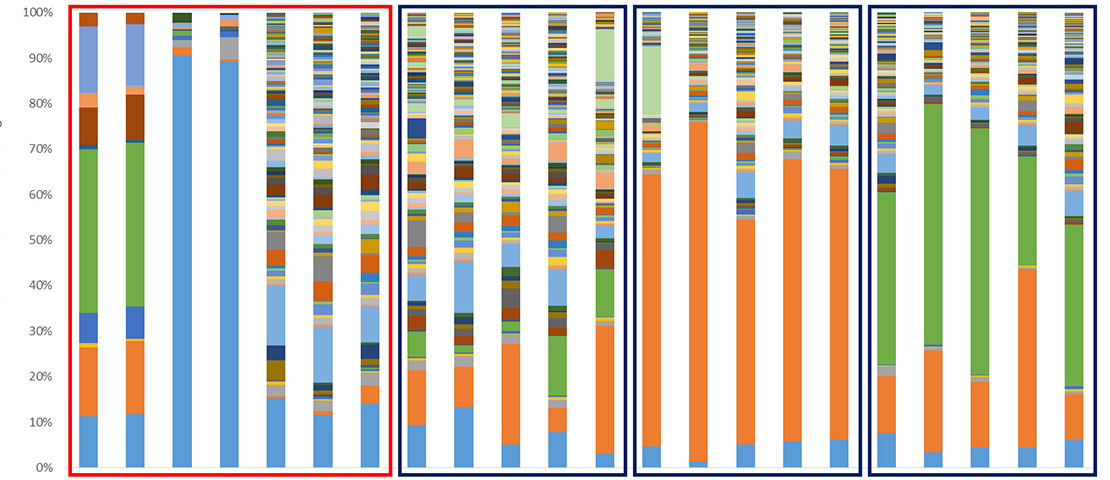
Microbial communities
Consisting in bacteria, archaea, protists, fungi and/or viruses previously unknown, microbial or microbial communities are now recognized as key actors in the proper functioning of our organism and our environment. Since 2008, GenoScreen has a particular focus on the study of these microbial communities and its R&D team has developed, optimized and standardized various methodologies mandatory for their study (Metabiote®, WHORMSS® etc), starting with the extraction of gDNA adapted to different samples of human/animal microbiotes (faeces, skin samples, oral, sputum, intestinal biopsies etc.) or environmental microbiotes (agricultural/polluted soils, rhizospheres, filtered air etc.) to the final metadata analysis.