Deeplex® Myc-TB is an all-in-one test for species-level identification, genotyping and prediction of antibiotic resistance in Mycobacterium tuberculosis complex strains. As the first ever tool to exploit in-depth DNA sequencing technology and thus detect and analyze antibiotic resistance, Deeplex® Myc-TB detects mutations responsible of antituberculosis drugs resistance.
A rapid and precise characterization
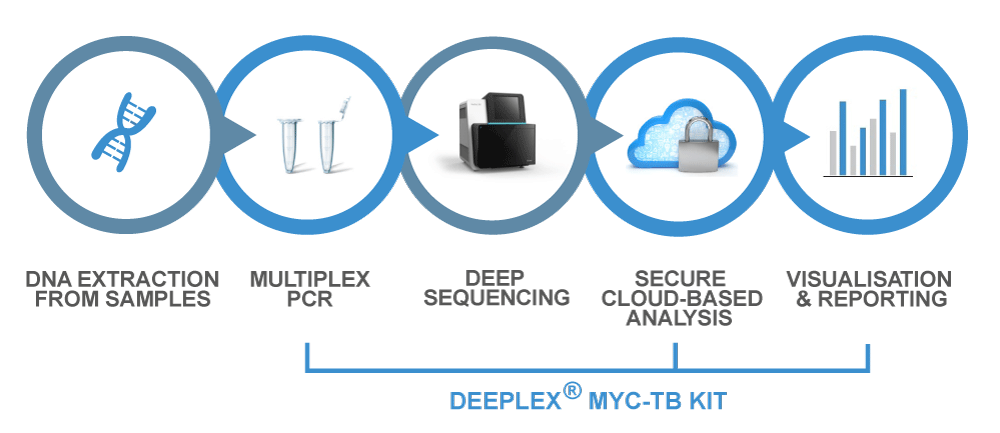
Based on next-generation sequencing, Deeplex® Myc-TB includes a targeted amplification kit that can be used directly on clinical samples, with no need for a culture step, in less than 48 hours.
Deeplex® Myc-TB also provides typing information on the strain involved which is useful for epidemiological surveillance and facilitates the identification of other bacterial pathogens from the same genus.
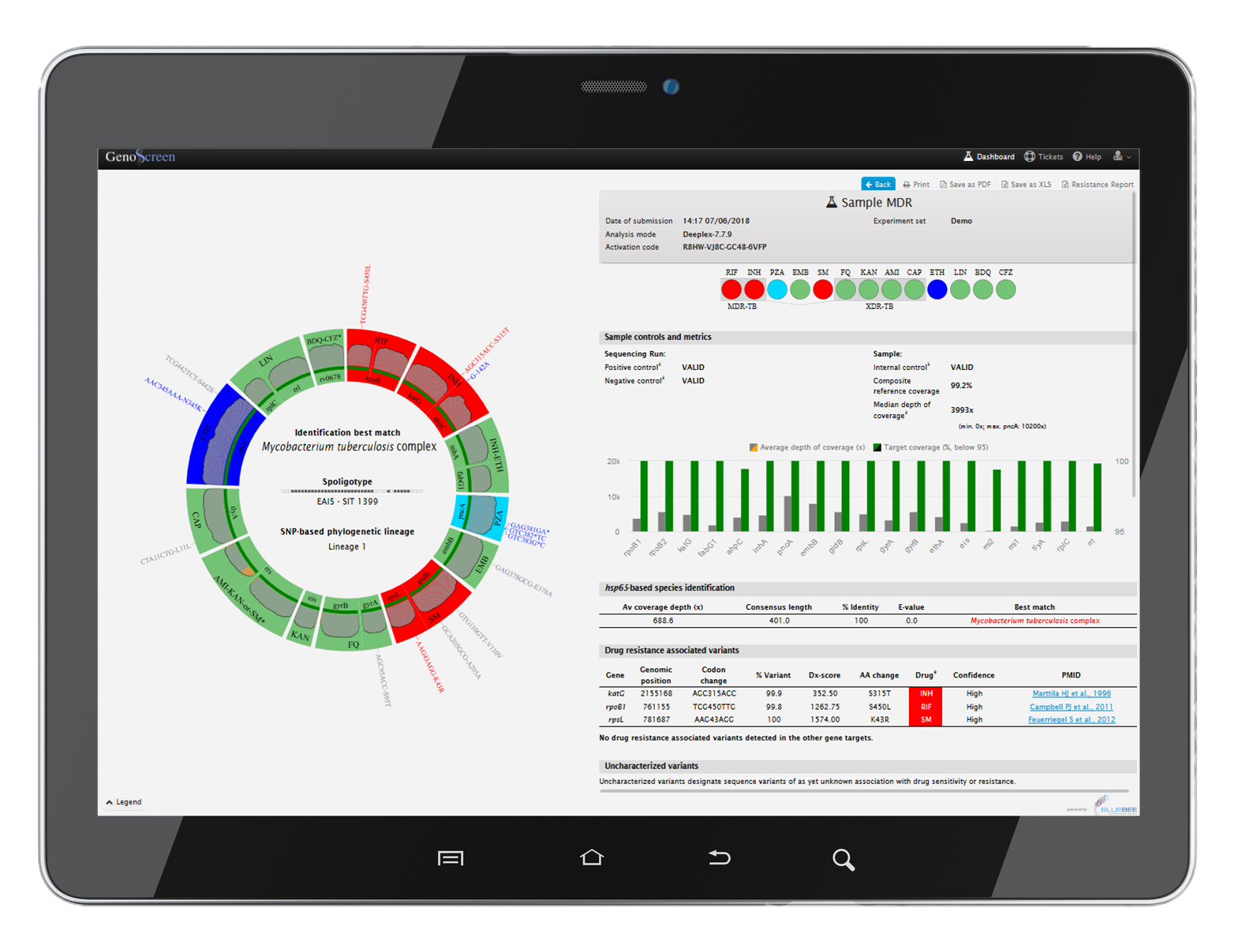
Deeplex® Myc-TB also includes an easy-to-use web application (hosted on a secure cloud digital platform) for the analysis and rapid interpretation of the results.
GenoScreen also commercializes other exclusive kits and services worldwide for the identification and typing of tuberculosis strains via the internationally standardized MIRU-VNTR. These solutions are used in epidemiological surveillance and for the detection of tuberculosis epidemics or other events (such as patient reinfections and laboratory contaminations). They are backed up by high-precision genome analysis services.
Deeplex® Myc-TB Workflow
- Directly applicable to clinical samples
- Simultaneous prediction of resistance to 15 anti-TB drugs
- MTBC hetero-resistance down to 3%
- MTBC spoligotyping and genotyping
- Identification of more than 100 mycobacterial species (NTM)
- Quick turnaround time of about 48 hours
- Multiplexing of up to 384 samples
Download the technical note:
Deeplex® is registered as a trademark in the USA (number 4980234) and in other countries.
The design of the visualization of the results in the Deeplex® web application is registered in the USA (number 890,770) and in other countries.