By applying a broad panel of genomic technologies and services, GenoScreen provides various solutions in identifying microorganisms via DNA sequencing and comparison with international databases.
These powerful and precised solutions are able to identify and characterize the microorganisms of interest: the species, the sub-species or the strain, typing, tracking and antibiotic resistance.
Identification and characterization
In order to identify a specie, ribosomal DNA regions targeted are adapted to each microorganism.
Additionally, to refine and adapt the identification to your needs, other genomic targets (rpoB and gyrB genes, etc.) are then used (discrimination within a group of species sharing the same ribosomal DNA sequence, for example).
GenoScreen can also leverage its know-how in whole-genome sequencing for a more precised characterization of a strain : virulence, antibiotic resistance, genomic comparisons with the strain library, etc.
Microbial typing and tracking
Our solution precisely characterizes any microorganism at the sub-species or strain level by using standardized, validated molecular methods, such as multilocus sequence typing (MLST) and multiple loci VNTR analysis (MLVA).
The typing schemes in open-access databases (PubMLST, MLST.org and MicrobesGenotyping) are used as references for these analyses.
The analysis can also be performed on a custom typing scheme.
GenoScreen, the world leader in tuberculosis genotyping
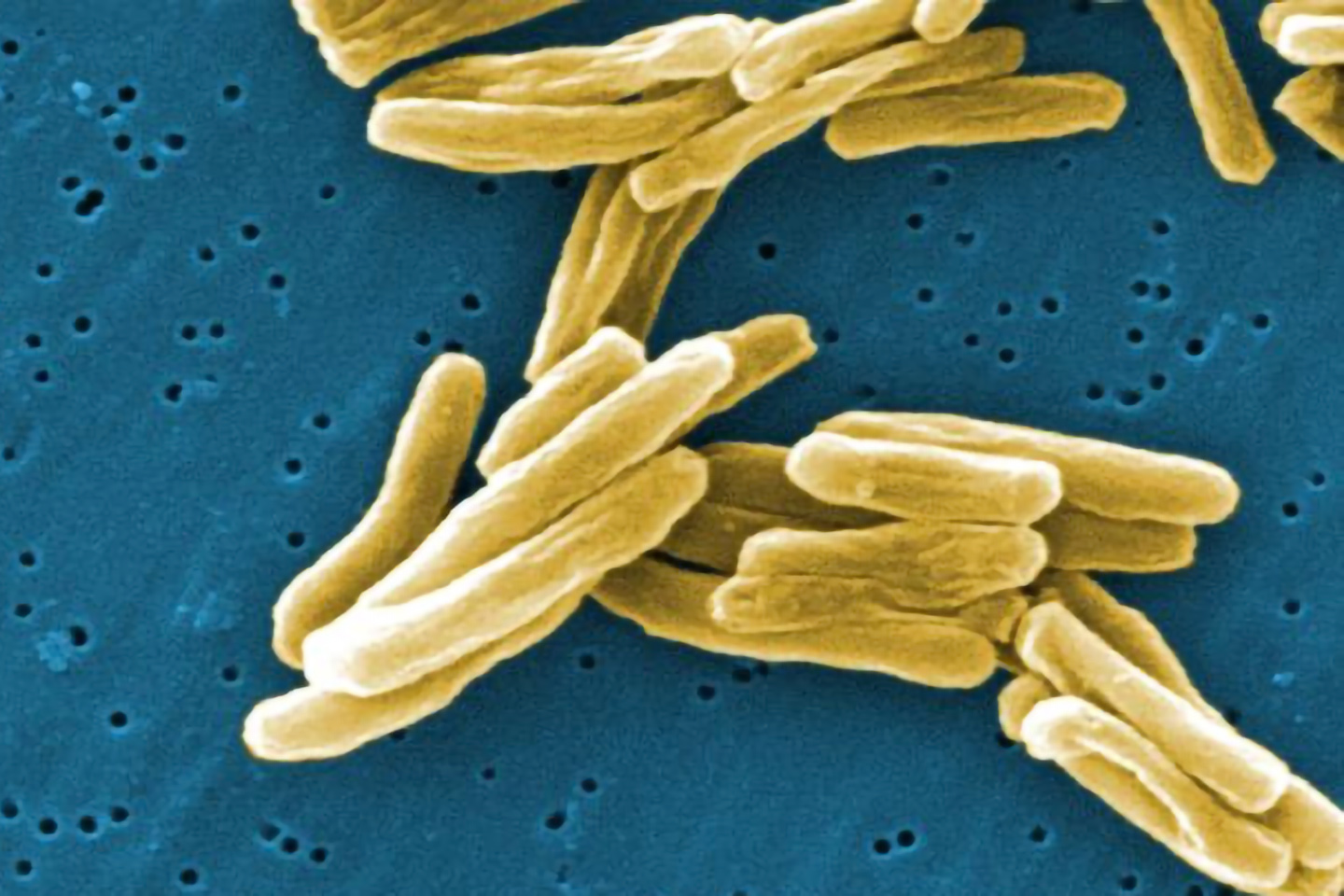
Credits to its cutting-edge technological know-how, GenoScreen is now the leader worldwide in genotyping and tracking of the Mycobacterium tuberculosis complex strains (the agent that causes tuberculosis).
The MIRU-VNTR solution developed by our teams is used as a kit or a service by many healthcare centers and research groups. A complementary spoligotyping approach is also available for identifying sub-species.
We propose kits and training courses for the implementation and an optimized use of the MIRU-VNTR method on Applied Biosystems® and QIAGEN® sequencers in your hospital or lab.
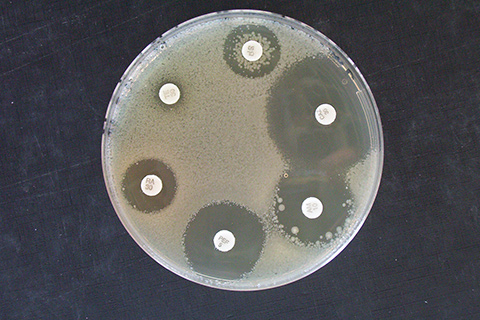
Antibiotic resistance
GenoScreen determines antibiotic resistance profiles via targeted sequencing of genes known to be associated with antibiotic resistance or by whole-genome sequencing. The sequences are compared with open-access databases or the literature data.
In-depth NGS sequencing can detect the presence of low-frequency variants conferring resistance (heteroresistance).
Deeplex®-MycTB - The all-in-one solution for characterizing Mycobacterium tuberculosis
The Deeplex®-MycTB solution developed by GenoScreen is a major innovation for the identification, characterization and epidemiological surveillance of tuberculosis for research purposes.
Deeplex®-MycTB is an all-in-one test for species-level identification, genotyping and prediction of antibiotic resistance in Mycobacterium tuberculosis complex strains.