Tuberculosis :
Predict in 48 hours the resistance
to the 15 major antibiotics
News
-
How to preserve the skin microbiota?
The skin microbiota plays a vital role in maintaining the skin's integrity and functionality. It contributes to the proper regulation of organisms and organs, allowing the skin to perform its essential functions, including acting as a barrier. See our latest article "Why preservce the skin microbiota ?"
-
GenoScreen bioinformatics training courses 2024
As a training organization, GenoScreen is offering bioinformatics training courses for professionals in 2024. Whether you are a novice or an expert, our training courses will give you the skills you need to master the analysis of NGS data and gain autonomy.
-
Why preserve the skin microbiota?
The skin serves as the first line of interaction with the external environment and as a defence mechanism against the physical, chemical, or biological stresses and challenges it encounters. The skin microbiota plays a key role in these interactions and defence functions. Formerly known as "skin flora", it colonises every square centimetre of the skin. Its role is fundamental to both skin health and the overall well-being of the body. What are the factors that influence its composition, and what disrupts its...
Focus
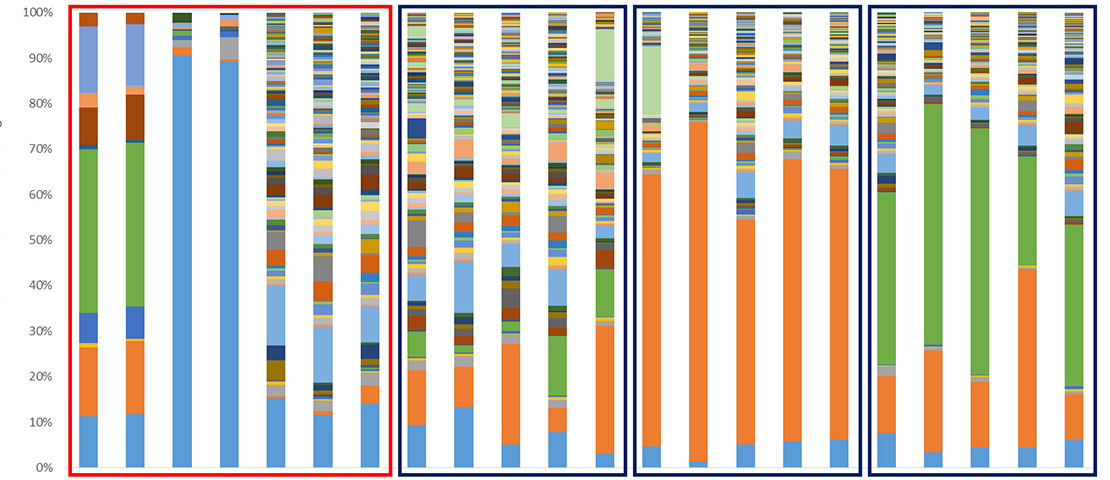
Microbial communities
Consisting in bacteria, archaea, protists, fungi and/or viruses previously unknown, microbial or microbial communities are now recognized as key actors in the proper functioning of our organism and our environment. Since 2008, GenoScreen has a particular focus on the study of these microbial communities and its R&D team has developed, optimized and standardized various methodologies mandatory for their study (Metabiote®, WHORMSS® etc), starting with the extraction of gDNA adapted to different samples of human/animal microbiotes (faeces, skin samples, oral, sputum, intestinal biopsies etc.) or environmental microbiotes (agricultural/polluted soils, rhizospheres, filtered air etc.) to the final metadata analysis.